Research results
Highlights

The Cell Tracking Challenge: 10 years of objective benchmarking
Maška, M., Ulman, V., Delgado-Rodriguez, P., Gómez-De-Mariscal, E., Nečasová, T., Peña, F. a. G., Ren, T. I., Meyerowitz, E. M., Scherr, T., Löffler, K., et al. (2023).
DOI: 10.1038/s41592-023-01879-y

Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4
Skrott, Z., Mistrik, M., Kaae Andersen, K., Friis, S., Majera, D., Gursky, J., Ozdian, T., Bartkova, J., Turi, Z., Moudry, P., Kraus, M., Michalova, et al. (2017).
DOI: 10.1038/nature25016
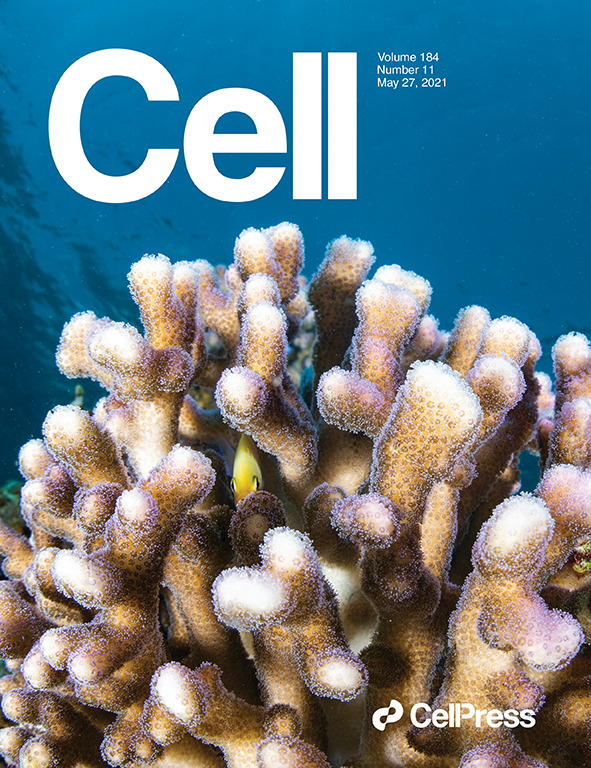
A cellular and spatial map of the choroid plexus across brain ventricles and ages
Dani, N; Herbst, RH; McCabe, C; Green, GS; Kaiser, K; Head, JP; Cui, J; Shipley, FB; Jang, A; Dionne, D; Nguyen, L; Rodman, C; Riesenfeld, SJ; Prochazka, et al. (2021).
DOI: 10.1016/j.cell.2021.04.003

Flagellar microtubule doublet assembly in vitro reveals a regulatory role of tubulin C-terminal tails
Schmidt-Cernohorska M, Zhernov I, Steib E, Le Guennec M, Achek R, Borgers S, Demurtas D, et al. (2019).
DOI: 10.1126/science.aav2567

ABP1-TMK auxin perception for global phosphorylation and auxin canalization
Friml, J; Gallei, M; Gelova, Z; Johnson, A; Mazur, E; Monzer, A; Rodriguez, L; Roosjen, M; Verstraeten, I; Zivanovic, BD; Zou, MX; Fiedler, et al. (2022).
DOI: 10.1038/s41586-022-05187-x

Autoimmune amelogenesis imperfecta in patients with APS-1 and coeliac disease
Gruper, Y., Wolff, A. S. B., Glanz, L., Spoutil, F., Marthinussen, M. C., Osičková, A., Herzig, Y., Goldfarb, Y., Aranaz‐Novaliches, G., Dobeš, J., et al. (2023).
DOI: 10.1038/s41586-023-06776-0
List of top publications
Fabbri, R., Scidà, A., Saracino, E., Conte, G., Kovtun, A., Candini, A., Kirdajova, D., Spennato, D., Marchetti, V., Lazzarini, C., Konstantoulaki, A., Dambruoso, P., Caprini, M., Muccini, M., Ursino, M., Anderova, M., Treossi, E., Zamboni, R., Palermo, V., & Benfenati, V. (2024). Graphene oxide electrodes enable electrical stimulation of distinct calcium signalling in brain astrocytes. Nature Nanotechnology, 19(2), 149–159.
DOI: 10.1038/s41565-024-01711-4
IF: 38.1
Dragwidge, J. M., Wang, Y. N., Brocard, L., De Meyer, A., Hudecek, R., Eeckhout, D., Grones, P., Buridan, M., Chambaud, C., Pejchar, P., Potocky, M., Winkler, J., Vandorpe, M., Serre, N., Fendrych, M., Bernard, A., De Jaeger, G., Pleskot, R., Fang, X. F., Van Damme, D., & Brocard, L. (2024). Biomolecular condensation orchestrates clathrin-mediated endocytosis in plants. Nature Cell Biology, 26(1), 1–13.
DOI: 10.1038/s41556-024-01354-6
IF: 17.3
Luo, Y., Lenarčič Živkovič, M., Wang, J., Ryneš, J., Foldynová-Trantírková, S., & Trantírek, L. (2024). A sodium/potassium switch for G4-prone G/C-rich sequences. Nucleic Acids Research, 52(1), 1–13.
DOI: 10.1093/nar/gkad1073
IF: 16.6
Palek, M., Palkova, N., Kleiblova, P., Kleibl, Z., & Macurek, L. (2024). RAD18 directs DNA double-strand break repair by homologous recombination to post-replicative chromatin. Nucleic Acids Research, 52(3), 1491–1504.
DOI: 10.1093/nar/gkae499
IF: 16.6
Hrychova, K., Burdova, K., Polackova, Z., Giamaki, D., Valtorta, B., Brazina, J., Krejcikova, K., Kuttichova, B., Caldecott, K. W., & Hanzlikova, H. (2024). Dispensability of HPF1 for cellular removal of DNA single-strand breaks. Nucleic Acids Research, 52(4), 1971–1985.
DOI: 10.1093/nar/gkae708
IF: 16.6
Codispoti, S., Yamaguchi, T., Makarov, M., Giacobelli, V. G., Masek, M., Kolár, M. H., Rocha, A. C. S., Fujishima, K., Zanchetta, G., & Hlouchová, K. (2024). The interplay between peptides and RNA is critical for protoribosome compartmentalization and stability. Nucleic Acids Research, 52(5), 1234–1247.
DOI: 10.1093/nar/gkae823
IF: 16.6
Visková, P., Istvánková, E., Rynes, J., Dzatko, S., Loja, T., Zivkovic, M. L., Rigo, R., El-Khoury, R., Serrano-Chacón, I., Damha, M. J., González, C., Mergny, J. L., Foldynová-Trantírková, S., & Trantírek, L. (2024). In-cell NMR suggests that DNA i-motif levels are strongly depleted in living human cells. Nature Communications, 15(1), 5401.
DOI: 10.1038/s41467-024-46221-y
IF: 14.7
Bulvas, O., Knejzlik, Z., Sys, J., Filimonenko, A., Cizkova, M., Clarová, K., Rejman, D., Kouba, T., & Pichová, I. (2024). Deciphering the allosteric regulation of mycobacterial inosine-5′-monophosphate dehydrogenase. Nature Communications, 15(2), 5510.
DOI: 10.1038/s41467-024-50933-6
IF: 14.7
Machacova, Z., Chroma, K., Lukac, D., Protivankova, I., & Moudry, P. (2024). DNA polymerase α-primase facilitates PARP inhibitor-induced fork acceleration and protects BRCA1-deficient cells against ssDNA gaps. Nature Communications, 15(3), 6501.
DOI: 10.1038/s41467-024-51667-1
IF: 14.7
Koval, T., Borah, N., Sudzinová, P., Brezovská, B., Sanderová, H., Hausnerová, V. V., Krenková, A., Hubálek, M., Trundová, M., Adamková, K., Dusková, J., Schwarz, M., Wiedermannová, J., Dohnálek, J., Krásny, L., & Kouba, T. (2024). Mycobacterial HelD connects RNA polymerase recycling with transcription initiation. Nature Communications, 15(4), 8701.
DOI: 10.1038/s41467-024-52891-5
IF: 14.7
Kachale, A., Pavlíková, Z., Nenarokova, A., Roithová, A., Durante, I. M., Miletínová, P., Záhonová, K., Nenarokov, S., Votýpka, J., Horáková, E., Ross, R. L., Yurchenko, V., Beznosková, P., Paris, Z., Valášek, L. S., & Lukeš, J. (2023). Author Correction: Short tRNA anticodon stem and mutant eRF1 allow stop codon reassignment. Nature, 618(7965), E23.
DOI: 10.1038/s41586-023-06260-9
IF 64,80
Gruper, Y., Wolff, A. S. B., Glanz, L., Spoutil, F., Marthinussen, M. C., Osičková, A., Herzig, Y., Goldfarb, Y., Aranaz‐Novaliches, G., Dobeš, J., Kadouri, N., Ben-Nun, O., Binyamin, A., Lavi, B., Givony, T., Khalaila, R., Gome, T., Wald, T., Mrázková, B., . . . Abramson, J. (2023). Autoimmune amelogenesis imperfecta in patients with APS-1 and coeliac disease. Nature, 624(7992), 653–662.
DOI: 10.1038/s41586-023-06776-0
IF 64,80
Maška, M., Ulman, V., Delgado-Rodriguez, P., Gómez-De-Mariscal, E., Nečasová, T., Peña, F. a. G., Ren, T. I., Meyerowitz, E. M., Scherr, T., Löffler, K., Mikut, R., Guo, T., Wang, Y., Allebach, J. P., Bao, R., Al-Shakarji, N. M., Rahmon, G., Toubal, I. E., Palaniappan, K., . . . Ortiz‐de‐Solórzano, C. (2023). The Cell Tracking Challenge: 10 years of objective benchmarking. Nature Methods, 20(7), 1010–1020.
DOI: 10.1038/s41592-023-01879-y
IF 47,99
Storchová, R., Palek, M., Palkova, N., Veverka, P., Brom, T., Hofr, C., & Macůrek, L. (2023). Phosphorylation of TRF2 promotes its interaction with TIN2 and regulates DNA damage response at telomeres. Nucleic Acids Research, 51(3), 1154–1172.
DOI: 10.1093/nar/gkac1269
IF 19,16
Fulneček, J., Klimentová, E., Cairó, A., Bukovcakova, S. V., Alexiou, P., Prokop, Z., & Říha, K. (2023b). The SAP domain of Ku facilitates its efficient loading onto DNA ends. Nucleic Acids Research, 51(21), 11706–11716.
DOI: 10.1093/nar/gkad850
IF 19,16
Martinek, J., Cifrová, P., Vosolsobě, S., García-González, J., Malínská, K., Mauerová, Z., Jelínková, B., Krtková, J., Sikorová, L., Leaves, I., Sparkes, I., & Schwarzerová, K. (2023). ARP2/3 complex associates with peroxisomes to participate in pexophagy in plants. Nature Plants, 9(11), 1874–1889.
DOI: 10.1038/s41477-023-01542-6
IF 17,35
Stibůrek, M., Ondráčková, P., Tučková, T., Turtaev, S., Šiler, M., Pikálek, T., Jákl, P., Gomes, A. D., Krejčí, J., Kolbábková, P., Uhlı́Řová, H., & Čižmár, T. (2023). 110 μm thin endo-microscope for deep-brain in vivo observations of neuronal connectivity, activity and blood flow dynamics. Nature Communications, 14(1).
DOI: 10.1038/s41467-023-36889-z
IF 16,60
Bohuslavová, R., Fabriciova, V., Smolík, O., Lebrón-Mora, L., Abaffy, P., Benesova, S., Žucha, D., Valihrach, L., Berková, Z., Saudek, F., & Pavlínková, G. (2023). NEUROD1 reinforces endocrine cell fate acquisition in pancreatic development. Nature Communications, 14(1).
DOI: 10.1038/s41467-023-41306-6
IF 16,60
Marečková, K., Mareček, R., Jáni, M., Žáčková, L., Andrýsková, L., Brázdil, M., & Nikolova, Y. S. (2023). Association of maternal depression during pregnancy and recent stress with brain age among adult offspring. JAMA Network Open, 6(1), e2254581.
DOI: 10.1001/jamanetworkopen.2022.54581
IF 13,80
Friml, J; Gallei, M; Gelova, Z; Johnson, A; Mazur, E; Monzer, A; Rodriguez, L; Roosjen, M; Verstraeten, I; Zivanovic, BD; Zou, MX; Fiedler, L; Giannini, C; Grones, P; Hrtyan, M; Kaufmann, WA; Kuhn, A; Narasimhan, M; Randuch, M; Rydza, N; Takahashi, K; Tan, ST; Teplova, A; Kinoshita, T; Weijers, D; Rakusova, H. ABP1-TMK auxin perception for global phosphorylation and auxin canalization. Nature.
DOI: 10.1038/s41586-022-05187-x
IF 69,50
Cairo, A; Vargova, A; Shukla, N; Capitao, C; Mikulkova, P; Valuchova, S; Pecinkova, J; Bulankova, P; Riha, K. Meiotic exit in Arabidopsis is driven by P-body-mediated inhibition of translation. Science.
DOI: 10.1126/science.abo0904
IF 63,70
Knizkova, D; Pribikova, M; Draberova, H; Semberova, T; Trivic, T; Synackova, A; Ujevic, A; Stefanovic, J; Drobek, A; Huranova, M; Niederlova, V; Tsyklauri, O; Neuwirth, A; Tureckova, J; Stepanek, O; Draber, P. CMTM4 is a subunit of the IL-17 receptor and mediates autoimmune pathology. Nat. Immunol.
DOI: 10.1038/s41590-022-01325-9
IF 31,25
Fry, M. Y., Najdrová, V., Maggiolo, A. O., Saladi, S. M., Doležal, P., & Clemons, W. M. Structurally derived universal mechanism for the catalytic cycle of the tail-anchored targeting factor Get3. Nat Struct Mol Biol.
DOI: 10.1038/s41594-022-00798-4
IF 18,36
Vaitsiankova, A; Burdova, K; Sobol, M; Gautam, A; Benada, O; Hanzlikova, H; Caldecott, KW. PARP inhibition impedes the maturation of nascent DNA strands during DNA replication. Nat. Struct. Mol. Biol.
DOI: 10.1038/s41594-022-00747-1
IF 18,36
Cihlarova, Z; Kubovciak, J; Sobol, M; Krejcikova, K; Sachova, J; Kolar, M; Stanek, D; Barinka, C; Yoon, G; Caldecott, KW; Hanzlikova, H. BRAT1 links Integrator and defective RNA processing with neurodegeneration. Nat. Commun.
DOI: 10.1038/s41467-022-32763-6
IF 17,69
Honek, J; Sramek, M; Honek, T; Tomek, A; Sefc, L; Januska, J; Fiedler, J; Horvath, M; Novotny, S; Brabec, M; Veselka, J. Screening and Risk Stratification Strategy Reduced Decompression Sickness Occurrence in Divers With Patent Foramen Ovale. JACC-Cardiovasc. Imag.
DOI: 10.1016/j.jcmg.2021.06.019
IF 16,05
Kopejtka K.,Tomasch J.,Kaftan D.,Gardiner T.A.,Bína D.,Gardian Z.,Bellas Ch.,Dröge A.,Geffers R.,Sommaruga R.,Koblížek M. A bacterium from a mountain lake harvests light using both proton-pumping xanthorhodopsins and bacteriochlorophyll-based photosystems. PNAS
DOI: 10.1073/pnas.2211018119
IF 12,77
Kashkan, I; Hrtyan, M; Retzer, K; Humpolickova, J; Jayasree, A; Filepova, R; Vondrakova, Z; Simon, S; Rombaut, D; Jacobs, TB; Frilander, MJ; Hejatko, J; Friml, J; Petrasek, J; Ruzicka, K. Mutually opposing activity of PIN7 splicing isoforms is required for auxin-mediated tropic responses in Arabidopsis thaliana. New Phytol.
DOI: 10.1111/nph.17792
IF 10,15
Škrott Zdeněk; Mistrík Martin; Hajdúch Marián; Džubák Petr; Bártek Jiří; Zbořil Radek. Bioavailable Dithiocarbamatemetal Complex Nanoparticles, Method of Preparation and Use Thereof. Patent: CA 3,076,855
Dani, N; Herbst, RH; McCabe, C; Green, GS; Kaiser, K; Head, JP; Cui, J; Shipley, FB; Jang, A; Dionne, D; Nguyen, L; Rodman, C; Riesenfeld, SJ; Prochazka, J; Prochazkova, M; Sedlacek, R; Zhang, F; Bryja, V; Rozenblatt-Rosen, O; Habib, N; Regev, A; Lehtinen, MK. A cellular and spatial map of the choroid plexus across brain ventricles and ages. Cell.
DOI: 10.1016/j.cell.2021.04.003
IF 41,50
Loubalova, Z; Fulka, H; Horvat, F; Pasulka, J; Malik, R; Hirose, M; Ogura, A; Svoboda, P. Formation of spermatogonia and fertile oocytes in golden hamsters requires piRNAs. Nat. Cell Biol.
DOI: 10.1038/s41556-021-00746-2
IF 28,82
Seda, V; Vojackova, E; Ondrisova, L; Kostalova, L; Sharma, S; Loja, T; Pavlasova, GM; Zicha, D; Peskova, MK; Krivanek, J; Liskova, K; Kren, L; Benes, V; Litzmanova, KM; Borsky, M; Oppelt, J; Verner, J; Pospisilova, S; Brychtova, Y; Panovska, A; Tan, Z; Zhang, SX; Doubek, M; Cerna, KA; Mayer, J; Mraz, M. FoxO1-GAB1 axis regulates homing capacity and tonic AKT activity in chronic lymphocytic leukemia. Blood.
DOI: 10.1182/blood.2020008101
IF 23,63
Horváthová, L.; Žárský, V.; Pánek, T.; Derelle, R.; Pyrih, J.; Motyčková, A.; Klápšťová, V.; Vinopalová, M.; Marková, L.; Voleman, L.; Klimeš, V.; Petrů, M.; Vaitová, Z.; Čepička, I.; Hryzáková, K.; Harant, K.; Gray, M. W.; Chami, M.; Guilvout, I.; Francetic, O.; Franz Lang, B.; Vlček, Č.; Tsaousis, A.D.; Doležal, P. Analysis of diverse eukaryotes suggests the existence of an ancestral mitochondrial apparatus derived from the bacterial type II secretion system. Nature Communications.
DOI: 10.1038/s41467-021-23046-7
IF 14,92
Mistrik, M; Skrott, Z; Muller, P; Panacek, A; Hochvaldova, L; Chroma, K; Buchtova, T; Vandova, V; Kvitek, L; Bartek, J. Microthermal-induced subcellular-targeted protein damage in cells on plasmonic nanosilver-modified surfaces evokes a two-phase HSP-p97/VCP response. Nature Communications.
DOI: 10.1038/s41467-021-20989-9
IF 14,92
Savkova, K; Huszar, S; Barath, P; Pakanova, Z; Kozmon, S; Vancova, M; Tesarova, M; Blasko, J; Kalinak, M; Singh, V; Kordulakova, J; Mikusova, K. An ABC transporter Wzm-Wzt catalyzes translocation of lipid-linked galactan across the plasma membrane in mycobacteria. Proc. Natl. Acad. Sci. U. S. A.
DOI: 10.1073/pnas.2023663118
IF 11,21
Synek, L; Pleskot, R; Sekeres, J; Serrano, N; Vukasinovic, N; Ortmannova, J; Klejchova, M; Pejchar, P; Batystova, K; Gutkowska, M; Jankova-Drdova, E; Markovic, V; Pecenkova, T; Santrucek, J; Zarsky, V; Potocky, M. Plasma membrane phospholipid signature recruits the plant exocyst complex via the EX070A1 subunit. Proc. Natl. Acad. Sci. U. S. A.
DOI: 10.1073/pnas.2105287118
IF 11,21
Lamos, M; Moravkova, I; Ondracek, D; Bockova, M; Rektorova, I. Altered Spatiotemporal Dynamics of the Resting Brain in Mild Cognitive Impairment with Lewy Bodies. Mov. Disord.
DOI: 10.1002/mds.28741
IF 10,34
Ezrova, Z; Nahacka, Z; Stursa, J; Werner, L; Vlcak, E; Viziova, PK; Berridge, MV; Sedlacek, R; Zobalova, R; Rohlena, J; Boukalova, S; Neuzil, J. SMAD4 loss limits the vulnerability of pancreatic cancer cells to complex I inhibition via promotion of mitophagy. Oncogene.
DOI: 10.1038/s41388-021-01726-4
IF 9,87
Albuquerque, LJC; Sincari, V; Jager, A; Kucka, J; Humajova, J; Pankrac, J; Paral, P; Heizer, T; Janouskova, O; Davidovich, I; Talmon, Y; Pouckova, P; Stepanek, P; Sefc, L; Hruby, M; Giacomelli, FC; Jager, E. pH-responsive polymersome-mediated delivery of doxorubicin into tumor sites enhances the therapeutic efficacy and reduces cardiotoxic effects. J. Control. Release.
DOI: 10.1016/j.jconrel.2021.03.013
IF 9,78
Honěk J, Šrámek M, Honěk T, Tomek A, Šefc L, Januška J, Fiedler J, Horváth M, Novotný Š and Veselka J. Patent Foramen Ovale Closure Is Effective in Divers Long-Term Results From the DIVE-PFO Registry. Journal of the American College of Cardiology. 2020, 76(9), 1149-1150. ISSN 0735-1097.
DOI: 10.1016/j.jacc.2020.06.072
IF 20,59
Eva Janisiw, Marilina Raices, Fabiola Balmir, Luis F. Paulin, Antoine Baudrimont, Arndt von Haeseler, Judith L. Yanowitz, Verena Jantsch & Nicola Silva. Poly(ADP‐ribose) glycohydrolase coordinates meiotic DNA double‐strand break induction and repair independent of its catalytic activity. Nature Commun. 2020 (11) 4869.
DOI: 10.1038/s41467-020-18693-1
IF 13,60
Tan S, Zhang X, Kong W, Yang XL, Molnár G, Vondráková Z, et al. (2020). The lipid code-dependent phosphoswitch PDK1–D6PK activates PIN-mediated auxin efflux in Arabidopsis. Nat. Plants.
DOI: 10.1038/s41477-020-0648-9
IF 13,25
Broft P, Dzatko S, Krafcikova M, Wacker A, Hänsel‐Hertsch R, Dötsch V, Trantirek L & Schwalbe H. (2020). In‐Cell NMR Spectroscopy of Functional Riboswitch Aptamers in Eukaryotic Cells. Angew. Chem. Int. Ed., 59, 2-11.
DOI: 10.1002/anie.202007184
IF 12,96
Hanzlikova H, Prokhorova E, Krejcikova K, Cihlarova Z, Kalasova I, Kubovciak J, Sachova J, Hailstone R, Brazina J, Ghosh S, Cirak S, Gleeson JG, AhelI and Caldecott KW (2020). “Pathogenic ARH3 mutations result in ADP-ribose chromatin scars during DNA strand break repair.” Nature Communications 11(1).
DOI: 10.1038/s41467-020-17069-9
IF 12,12
Henrichs V, Grycova L, Barinka C, et al. Mitochondria‐adaptor TRAK1 promotes kinesin‐1 driven transport in crowded environments. Nat Commun. 2020;11(1):3123.
DOI: 10.1038/s41467-020-16972-5
IF 12,12
Krivanek J, Soldatov RA, Kastriti ME, Chontorotzea T, Herdina AN, Petersen J, Szarowska B, Landova M, Kovar Matejova V, Izakovicova Holla L, Kuchler U, Vidovic Zdrilic I, Vijaykumar A, Balic A, Marangoni P, Klein OD, Neves VCM, Yianni V, Sharpe PT, Harkany T, Metscher BD, Bajénoff M, Mina M, Fried K, Kharchenko, PV, & Adameyko I. (2020). Dental cell type atlas reveals stem and differentiated cell types in mouse and human teeth. Nature communications, 11, 4816, 1-18.
DOI: 10.1038/s41467-020-18512-7
IF 12,12
Pospisil J, Vitovska D, Kofronova O, Muchova K, Sanderova H, Hubalek M, Sikova M, Modrak M, Benada O, Barak I and Krasny L (2020). “Bacterial nanotubes as a manifestation of cell death.” Nat Commun 11(1): 4963.
DOI: 10.1038/s41467-020-18800-2
IF 12,12
Venit T, Semesta K, Farrukh S, Endara-Coll M, Havalda R, Hozak P and Percipalle P (2020). “Nuclear myosin 1 activates p21 gene transcription in response to DNA damage through a chromatin-based mechanism.” Communications Biology 3(1).
DOI: 10.1038/s42003-020-0836-1
IF 12,12
Voboril M, Brabec T, Dobes J, Splichalova I, Brezina J, Cepkova A, Dobesova M, Aidarova A, Kubovciak J, Tsyklauri O, Stepanek O, Benes V, Sedlacek R, Klein L, Kolar M and Filipp D (2020). “Toll-like receptor signaling in thymic epithelium controls monocyte-derived dendritic cell recruitment and Treg generation.” Nature Communications 11(1).
DOI: 10.1038/s41467-020-16081-3
IF 12,12
Schmidt-Cernohorska M, Zhernov I, Steib E, Le Guennec M, Achek R, Borgers S, Demurtas D, Mouawad L, Lansky Z, Hamel V, Guichard P. Flagellar microtubule doublet assembly in vitro reveals a regulatory role of tubulin C-terminal tails. Science. 363(6424), 285–288 (2019).
DOI: 10.1126/science.aav2567
IF 41,06
Siahaan V, Krattenmacher J, Hyman AA, Diez S, Hernández-Vega A, Lansky Z, Braun M. Kinetically distinct phases of tau on microtubules regulate kinesin motors and severing enzymes. Nature Cell Biology (9):1086-1092 (2019).
DOI: 10.1038/s41556-019-0374-6
IF 17,73
Kotrbová A, Štěpka K, Maška M, Pálenik J, Ilkovics L, Klemová D, Kravec M, Hubatka F, Dave Z, Hampl A, Bryja V, Matula P, Pospíchalová V. TEM ExosomeAnalyzer: a computer-assisted software tool for quantitative evaluation of extracellular vesicles in transmission electron microscopy images. Journal of Extracellular Vesicles, 2019, 8(1):1-13. ISSN 2001-3078.
DOI: 10.1080/20013078.2018.1560808
IF 14,98
Karnkowska A, Treitli SC, Brzoň O, Novák L, Vacek V, Soukal P, Barlow LD, Herman EK, Pipaliya SV, Pánek T, Žihala D, Petrželková R, Butenko A, Eme L, Stairs CW, Roger AJ, Eliáš M, Dacks JB, Hampl V. The Oxymonad Genome Displays Canonical Eukaryotic Complexity in the Absence of a Mitochondrion. Mol. Biol. Evol. (2019). 36(10):2292-2312.
DOI: 10.1093/molbev/msz147
IF 14,80
Krafčíková M, Džatko Š, CaronC, Granzhan A, Fiala R, Loja T, Teulade-Fichou MP, Fessl T, Hansel-Hertsch R, Mergny JL, Foldynova-Trantirkova S, Trantírek L. Monitoring DNA-Ligand Interactions in Living Human Cells Using NMR Spectroscopy. Journal of the American Chemical Society, 2019, 141(34), 13281-13285.
DOI: 10.1021/jacs.9b03031
IF 14,70
Kaiser K, Gyllborg D, Prochazka J, Salašová A, Kompaníková P, Molina FL, Laguna-Goya R, Radaszkiewicz TW, Harnoš J, Prochazkova M, Potěšil D, Barker RA, Casado AG, Zdráhal Z, Sedlacek R, Arenas E, Villaescusa JC a Bryja V. WNT5A is transported via lipoprotein particles in the cerebrospinal fluid to regulate hindbrain morphogenesis. Nature Commun., 2019, 10(1), 1-15.
DOI: 10.1038/s41467-019-09298-4
IF 13,60
Bouchal, P.; Dvořák, P.; Babocký, J.; Bouchal Z.; Ligmajer, F.; Hrtoň M.; Křápek. V.; Faßbender, A.; Linden, S.; Chmelík, R.; Šikola, T, 2019: High-Resolution Quantitative Phase Imaging of Plasmonic Metasurfaces with Sensitivity down to a Single Nanoantenna. Nano Letters 19 (2),1242-1250.
DOI: 10.1021/acs.nanolett.8b04776
IF 11,24
Fajkus P, Peška V, Závodník M, Fojtová M, Fulnečková J, Dobias S, Kilar AM, Dvořáčková M, Zachová D, Nečasová I, Sims J, Sýkorová E, Fajkus J. Telomerase RNAs in land plants. Nucleic acids research, 2019, 47(18), 9842-9856.
DOI: 10.1093/nar/gkz695
IF 11,15
Jelínková, A., Malínská, K., and Petrášek, J. (2019). Using FM Dyes to Study Endomembranes and Their Dynamics in Plants and Cell Suspensions. In Plant Cell Morphogenesis. Methods Mol Biol. 1992:173–187.
DOI: 10.1007/978-1-4939-9469-4_11
IF 10,71
Bajzikova M, Kovarova J, Coelho AR, Boukalova S, Oh S, Rohlenova K, Svec D, Hubackova S, Endaya B, Judasova K, Bezawork-Geleta A, Kluckova K, Chatre L, Zobalova R, Novakova A, Vanova K, Ezrova Z, Maghzal GJ, Magalhaes Novais S, Olsinova M, Krobova L, An YJ, Davidova E, Nahacka Z, Sobol M, Cunha-Oliveira T, Sandoval-Acuna C, Strnad H, Zhang T, Huynh T, Serafim TL, Hozak P, Sardao VA, Koopman WJH, Ricchetti M, Oliveira PJ, Kolar F, Kubista M, Truksa J, Dvorakova-Hortova K, Pacak K, Gurlich R, Stocker R, Zhou Y, Berridge MV, Park S, Dong L, Rohlena J, Neuzil J. Reactivation of Dihydroorotate Dehydrogenase-Driven Pyrimidine Biosynthesis Restores Tumor Growth of Respiration-Deficient Cancer Cells. Cell Metab 2018; pii:S1550-4131(18)30646-6. [Epub ahead of print].
DOI: 10.1016/j.cmet.2018.10.014
IF 20,57
Jirouskova M, Nepomucka K, Oyman-Eyrilmez G, Kalendova A, Havelkova H, Sarnova L, Chalupsky K, Schuster B, Benada O, Miksatkova P, Kuchar M, Fabian O, Sedlacek R, Wiche G, Gregor M. Plectin controls biliary tree architecture and stability in cholestasis. J Hepatol. 2018; 68(5): 1006-1017.
DOI: 10.1016/j.jhep.2017.12.011
IF 15,04
Hanzlikova H, Kalasova I, Demin AA, Pennicott LE, Cihlarova Z, Caldecott KW. The Importance of Poly(ADP-Ribose) Polymerase as a Sensor of Unligated Okazaki Fragments during DNA Replication. Mol Cell 2018; 71(2): 319-331.e3.
DOI: 10.1016/j.molcel.2018.06.004
IF 14,25
Bezawork-Geleta A, Wen H, Dong L, Yan B, Vider J, Boukalova S, Krobova L, Vanova K, Zobalova R, Sobol M, Hozak P, Novais SM, Caisova V, Abaffy P, Naraine R, Pang Y, Zaw T, Zhang P, Sindelka R, Kubista M, Zuryn S, Molloy MP, Berridge MV, Pacak K, Rohlena J, Park S, Neuzil J. Alternative assembly of respiratory complex II connects energy stress to metabolic checkpoints. Nat Commun 2018; 9(1): 2221.
DOI: 10.1038/s41467-018-04603-z
IF 12,40
Dzatko S, Krafcikova M, Hänsel-Hertsch R, Fessl T, Fiala R, Loja T, Krafcik D, Mergny JL, Foldynova-Trantirkova S, Trantirek L. Evaluation of the Stability of DNA i-Motifs in the Nuclei of Living Mammalian Cells. Angew Chem Int Ed Engl. 2018; 57(8): 2165-2169.
DOI: 10.1002/anie.201712284
IF 12,1
Kazamia E, Sutak R, Paz-Yepes J, Dorrell RG, Jimenez Vieira FR, Mach J, Morrissey J, Leon S, Lam F, Pelletier E, Camadro JM, Bowler C, Lesuisse E. Endocytosis-mediated siderophore uptake as a strategy for Fe acquisition in diatoms. Sci Adv 2018; 4(5): eaar4536 (2018).
DOI: 10.1126/sciadv.aar4536
IF 11,51
Magiera MM, Bodakuntla S, Ziak J, Lacomme S, Sousa PM, Leboucher S, Hausrat TJ, Bosc C, Andrieux A, Kneussel M, Landry M, Calas A, Balastik M, Janke C. Excessive tubulin polyglutamylation causes neurodegeneration and perturbs neuronal transport. EMBO Journal 37 (23): e100440, 2018.
DOI: 10.15252/embj.2018100440
IF 10,56
Kaurov I., Vancová M., Schimanski B., Cadena L.R., Heller J., Bílý T., Potěšil D., Eichenberger C., Bruce H., Oeljeklaus S., Warscheid B., Zdráhal Z., Schneider A, Lukeš J., H. Hashimi H., 2018: The Diverged Trypanosome MICOS Complex as a Hub for Mitochondrial Cristae Shaping and Protein Import. Current Biology 28: 3393-3407, e5
DOI: 10.1016/j.cub.2018.09.008
IF 9,97
Platre MP, Noack LC, Doumane M, Bayle V, Simon MLA, Maneta-Peyret L, Fouillen L, Stanislas T, Armengot L, Pejchar P, et al. (2018). A Combinatorial Lipid Code Shapes the Electrostatic Landscape of Plant Endomembranes. Developmental Cell 45: 465–480.e11.
DOI: 10.1016/j.devcel.2018.04.011
IF 9,62
Kania U, Nodzyński T, Lu Q, Hicks GR, Nerinckx W, Mishev K, Peurois F, Cherfils J, De Rycke R, Grones P, Robert S, Russinova E, Friml J. The Inhibitor Endosidin 4 Targets SEC7 Domain‐Type ARF GTPase Exchange Factors and Interferes with Subcellular Trafficking in Eukaryotes. Plant Cell. 2018 Oct;30(10):2553‐2572.
DOI: 10.1105/tpc.18.00127
IF 8,63
Skrott, Z., Mistrik, M., Andersen, K. et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature 552, 194–199 (2017).
DOI: 10.1038/nature25016
IF 41,58
Ulman V, Maška M, Magnusson KEG, Ronneberger O, Haubold C, Harder N, Matula P, Matula P, Svoboda D, Radojevic M, Smal I, Rohr K, Jaldén J, Blau HM, Dzyubachyk O, Lelieveldt B, Xiao P, Li Y, Cho S, Dufour AC, Olivo-Marin J, Reyes-Aldasoro CC, Solis-Lemus JA, Bensch R, Brox T, Stegmaier J, Mikut R, Wolf S, Hamprecht FA, Esteves T, Quelhas P, Demirel Ö, Malmström L, Jug F, Tomancak P, Meijering E, Muñoz-Barrutia A, Kozubek M, Ortiz-de-Solorzano C. An objective comparison of cell-tracking algorithms. Nature Methods, 2017, vol. 14, No 12, p. 1141-1152. ISSN 1548-7091. 2017.
DOI: 10.1038/nmeth.4473
IF 26,92
Lukeš, T., Glatzová, D., Kvíčalová, Z. et al. Quantifying protein densities on cell membranes using super-resolution optical fluctuation imaging. Nat Commun 8, 1731 (2017).
DOI: 10.1038/s41467-017-01857-x
IF 12,35
Robert HS, Park C, Gutièrrez CL, Wójcikowska B, Pěnčík A, Novák O, Chen J, Grunewald W, Dresselhaus T, Friml J, Laux T. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat Plants. 2018; 4(8): 548-553.
DOI: 10.1038/s41477-018-0204-z
IF 11,47
Bacakova L, Zarubova J, Travnickova M, Musilkova J, Pajorova J, Slepicka P, Kasalkova NS, Svorcik V, Kolska Z, Motarjemi H, Molitor M. Stem cells: their source, potency and use in regenerative therapies with focus on adipose-derived stem cells – a review. Biotechnology Advances 2018; 36: 1111–1126.
DOI: 10.1016/j.biotechadv.2018.03.011
IF 11,45
Skalický T., Dobáková E., Wheeler R.J., Tesařová M., Flegontov P., Jirsová D., Votýpka J., Yurchenko V., Ayala F., Lukeš J. 2017: Extensive flagellar remodeling during the complex life cycle of Paratrypanosoma, an early-branching trypanosomatid. PNAS 114: 11757-11762.
DOI: 10.1073/pnas.1712311114
IF 10,36
Dachev, M., Bína, D., Sobotka, R., Moravcová, L., Gardian, Z., Kaftan, D., Šlouf, V., Fuciman, M., Polívka, T. & Koblížek, M. 2017: Unique double concentric ring organization of light harvesting complexes in Gemmatimonas phototrophica. PLoS Biology 15(12): e2003943.
DOI: 10.1371/journal.pbio.2003943
IF 9,53
Petrlova et al. Aggregation of thrombin-derived C-terminal fragments as a previously undisclosed host defense mechanism. PNAS. 114:E4213-E4222.
DOI: 10.1073/pnas.1619609114
IF 9,50
Anderkova, L., Barton, M., & Rektorova, I. (2017). Striato-cortical connections in Parkinson’s and Alzheimer’s diseases: Relation to cognition. Movement Disorders.
DOI: 10.1002/mds.26956
IF 8,68
Dong LF, Kovarova J, Bajzikova M, Bezawork-Geleta A, Svec D, Endaya B, Sachaphibulkij K, Coelho AR, Sebkova N, Ruzickova A, Tan AS, Kluckova K, Judasova K, Zamecnikova K, Rychtarcikova Z, Gopalan V, Andera L, Sobol M, Yan B, Pattnaik B, Bhatraju N, Truksa J, Stopka P, Hozak P, Lam AK, Sedlacek R, Oliveira PJ, Kubista M, Agrawal A, Dvorakova-Hortova K, Rohlena J, Berridge MV, Neuzil J. Horizontal transfer of whole mitochondria restores tumorigenic potential in mitochondrial DNA-deficient cancer cells. Elife. 2017 Feb 15;6:e22187. PMID: 28195532; PMCID: PMC5367896.
DOI: 10.7554/eLife.22187
IF 7,73
Tucek J, Sofer Z, Bousa D, Pumera M, Hola K, Mala A, Polakova K, Havrdova M, Cepe K, Tomanec O, Zboril R. Air-stable superparamagnetic metal nanoparticles entrapped in graphene oxide matrix. Nature Communications, 2016; 7: 12879.
DOI: 10.1038/ncomms12879
IF 11,33
Chroma K, Mistrik M, Moudry P, Gursky J, Liptay M, Strauss R, Skrott Z, Vrtel R, Bartkova J, Kramara J, Bartek J. Oncogene. 2017 Apr 27;36(17):2405‐2422. Epub 2016 Nov 14. Tumors overexpressing RNF168 show altered DNA repair and responses to genotoxic treatments, genomic instability and resistance to proteotoxic stress. Oncogene 36, 2405–2422 (2017)
DOI: 10.1038/onc.2016.392
IF 6,85
Mistrik M., Vesela E., Furst T., Hanzlikova H., Frydrych I., Gursky J, Majera D. & Bartek J. A novel quantitative photo‐manipulation technique. Sci Rep. 2016 Jan 18;6:19567. Cells and Stripes.
DOI: 10.1038/srep19567
IF 4,12
Kratochvila, J; Jirik, R; Bartos, M; Standara, M; Starcuk, Z; Taxt, T: Distributed capillary adiabatic tissue homogeneity model in parametric multi‐channel blind AIF estimation using DCE‐MRI. Magnetic Resonance in Medicine 75 (2016), 1355‐1365.
DOI: 10.1002/mrm.25619
IF 3,92
Laňková M, Humpolíčková J, Vosolsobě S, Cit Z, Lacek J, Čovan M, Čovanová M, Hof M, Petrášek J. Determination of Dynamics of Plant Plasma Membrane Proteins with Fluorescence Recovery and Raster Image Correlation Spectroscopy. Microsc Microanal. 2016 Apr;22(2):290-9.
DOI: 10.1017/S1431927616000568
IF 2,12






